

In some cases building a guide tree can take a long time since it requires making a pairwise alignment between each pair of sequences. When pairwise aligning profiles, mismatch costs are weighted proportional to the fraction of mismatching bases and gap introduction and gap extension costs are proportionally reduced at sites where the other profile contains some gaps. This can then be pairwise aligned to another sequence or alignment profile. The resulting alignment is placed in the folder containing the original sequences.Ī profile is a matrix of numbers representing the proportion of symbols (nucleotide or amino acid) at each position in an alignment. The number of times each sequence is re-aligned is determined by the refinement iterations option in the multiple alignment window. “Refining” an alignment involves removing sequences from the alignment one at a time, and then realigning the removed sequence to a “profile” of the remaining sequences.
#Geneious prime alignment plus#
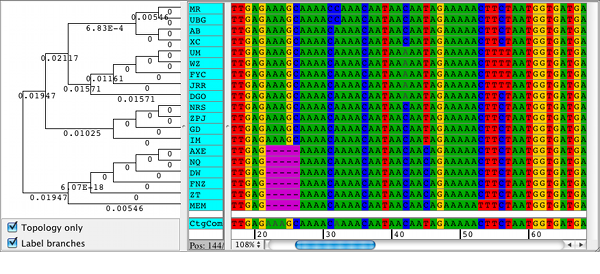
The neighbor-joining method of tree building is used to create the guide tree.Īs progressive pairwise alignment proceeds via a series of pairwise alignments, this function has all the standard pairwise alignment options, plus the option of refining the multiple sequence alignment once it is done. The Geneious multiple alignment algorithm uses progressive pairwise alignment. Select Geneious as the alignment algorithm. To run a multiple alignment in Geneious Prime, select all the sequences you wish to align and click Align/Assemble → Multiple align. Multiple sequence alignment in Geneious Prime
#Geneious prime alignment software#
Many variations of the progressive pairwise alignment algorithm exist, including the one used in the popular alignment software ClustalX. Their algorithm used a guide tree to choose which pair of sequences/alignments to align at each step. Feng & Doolittle were the first to describe progressive pairwise alignment. This process is repeated until a single alignment containing all of the sequences remains.

However a number of useful heuristic algorithms for multiple sequence alignment do exist. While this is an attractive option there are no efficient algorithms for doing this currently available.
One way to score such an alignment would be to use a probabilistic model of sequence evolution and select the alignment that is most probable given the model of evolution. Hence the need for automatic multiple sequence alignments based on objective criteria. Multiple sequence alignments can be done by hand but this requires expert knowledge of molecular sequence evolution and experience in the field. It is not always possible to clearly identify structurally or evolutionarily homologous positions and create a single “correct” multiple sequence alignment ( Durbin et al 1998). While handling protein sequences, it is important to be able to tell what a multiple sequence alignment means – both structurally and evolutionarily. It should be noted that protein sequences that are structurally very similar can be evolutionarily distant. A multiple sequence alignment can be used for many purposes including inferring the presence of ancestral relationships between the sequences. A multiple sequence alignment is a comparison of multiple related DNA or amino acid sequences.


 0 kommentar(er)
0 kommentar(er)
